Research Directions
Our lab is focusing on the following areas of research:
1) TGF? signaling and cancer
Transforming Growth Factor β (TGFβ) is a pleiotropic cytokine that controls
several key biological processes including cell proliferation, differentiation,
apoptosis and extracellular matrix production. TGFβ signals via heteromeric
complexes of type I and type II serine/threonine kinase receptors and
via intracellular effectors called Smads. Following their activation by
TGFβ, type I receptors phosphorylate a subset of the Smad family called
R-Smads (Smad2 and SMad3) causing their heterooligomerization with a common
Smad (Smad4) and their translocation to the nucleus where they activate
the transcription of target genes in cooperation with DNA binding cofactors
and coactivators.
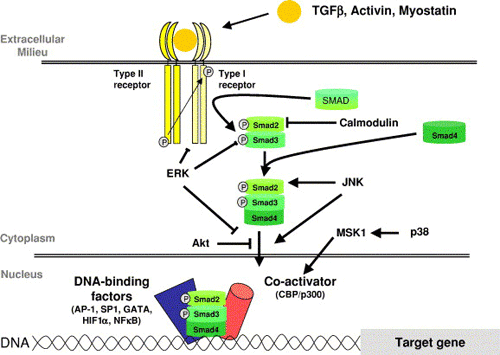
The basic TGF? signaling engine
Deregulation of TGFβ signaling either by mutations in key signaling components
or by negative interference with other signaling pathways is a common
characteristic in many types of cancer (pancreatic, colon, breast cancer).
We are concentrating our efforts on several aspects of TGFβ signaling.
We are studying the structure-function relationship in Smad proteins and
the characterization of tumorigenic mutations. By using a large panel
of Smad mutants generated in our laboratory, we are investigating the
structural requirements for Smad oligomerization, nuclear translocation
and transcriptional activation. We are characterizing in detail a novel
transactivation domain in the linker region of Smad3 identified recently
in our laboratory (Prokova et al, NAR 2005).Using various in vitro and
in vivo assays including a recently described protein-protein interaction
assay based on biotinylation in vivo, we are analyzing inactivating mutations
in Smad proteins found frequently in cancer patients. In collaboration
with Dr George Mosialos (Fleming Institute, Athens) and Dr Christos Tsatsanis
(U. of Crete Medical School) we are studying the molecular mechanisms
of inhibition of TGFβ signaling and Smad functions by oncogenic proteins
such as the Latent Membrane Protein 1 of the Epstein Barr Virus and the
product of the Tpl2/Cot oncogene. Finally, we are characterizing the mechanisms
of transcriptional activation by Smad proteins by analyzing the promoter
regions of novel TGFβ target genes. We have initiated this effort by focusing
on the promoter of the RhoB small GTPase gene which is involved in the
TGFβ-induced cytoskeletion reorganization in cancer cells (collaboration
with Dr Christos Stournaras, U. of Crete Medical School).
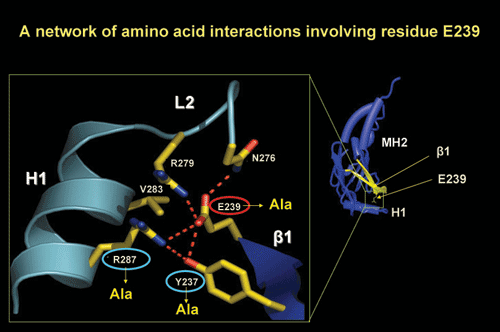
Structural detail in the region surrounding a tumorigenic mutation in
Smad4 protein (E239).
Mutations introduced are shown in yellow.
2) Regulation and functions of the cdk inhibitor
p21/WAF1
p21WAF1/Cip1 (p21) is a potent cell cycle inhibitor downstream of the
p53 tumor suppressor protein. In the nucleus, p21 is an inhibitor of G1
and G2 cyclin-dependent kinases as well as of the Proliferating Nuclear
Antigen (PCNA). In contrast, cytosolic sequestration of p21 which is a
frequent case in cancer cells, alters the functional properties of p21
from growth inhibitory to tumor promoting and metastatic. Promotion of
tumor growth and metastasis by cytosolic p21 is exerted via inhibition
of the apoptotic pathway and the regulation of actin cytoskeleton organization.
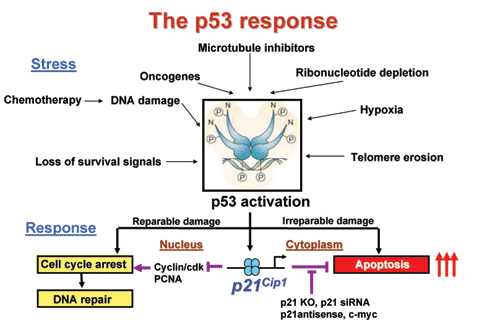
Summary of the various stress factors that active the p53 tumor suppressor
protein and the opposite functions played by the cdk inhibitor p21 in
nucleus and the cytoplasm.
We are interested in understanding the molecular mechanisms that govern
the response of the p21 gene to cytokines (TGFβ) and to chemotherapeutic
drugs that activate the p53 tumor suppressor protein (5-Fluorouracil,
Mithramycin A). We are studying the role played by members of the Sp1
family of ubiquitous transcription factors in the above p21 gene responses
by characterizing the physical and functional interactions between Sp
proteins and signaling mediators (p53, Smads, AP1).
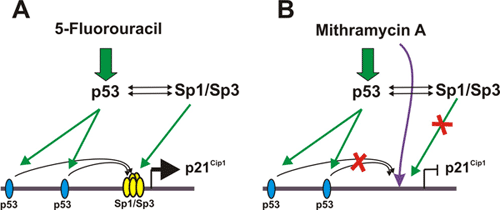
The role of transcription factor Sp1 in the transcriptional activation
of p21 gene by chemotherapeutic drugs.
As mentioned above, the intracellular localization of p21 determines the
decision between cell cycle arrest and apoptosis We are planning to apply
gene and protein transfer technologies in order to evaluate the therapeutic
potential of nuclear p21 in breast cancer. This will be accomplished by
delivering purified TAT-p21 fusion proteins or recombinant adenoviruses
expressing p21, either wild type or mutant, into breast cancer cells or
animals models of cancer and by following their growth and metastatic
properties. Furthermore, we are planning to characterize the regulatory
mechanisms involved in p21 nucleo-cytoplasmic shuttling in breast cancer
cells expressing high or low levels of the her/neu oncogene. We propose
to study the role of p21 in the apoptotic response of breast cancer cells
undergoing chemotherapy using gene silencing technology.
3) Transcriptional regulation of genes involved in lipoprotein
metabolism
a) Transcriptional regulation of the human apolipoprotein genes by hormore
nuclear receptors: For many years we have been studying, using in vitro
and in vivo approaches, the mechanisms that govern the regulation of transcription
of the genes coding for of all major human apolipoproteins in liver and
intestinal cells. In recent years we have been focusing our efforts on
the specific role of orphan and ligand dependent hormone nuclear receptors
on the expression of these genes by identifying and characterizing Hormone
Response Elements (HREs) present on the promoters and the enhancers of
the genes of the two apolipoprotein gene clusters in humans: the apoA-I/C-III/A-IV
gene cluster on chromosome 11 and the apoE/C-I/C-IV/C-II gene cluster
on chromosome 19. We are also studying the effect of pro- and inflammatory
cytokines on the transcriptional activity of huclear receptors and the
expression of genes of the apoA-I/C-III/A-IV gene cluster,
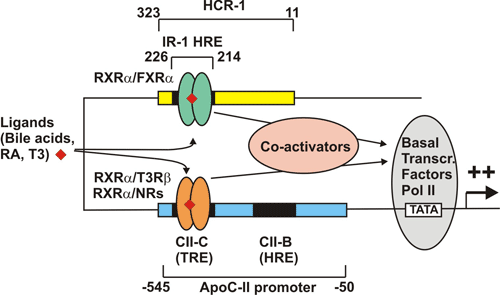
Schematic representation of putative synergistic interactions between
nuclear receptors bound to the IR-1 of the (214/226) HCR-1 and the HREs
CIIB and CIIC present in the proximal apoC-II promoter
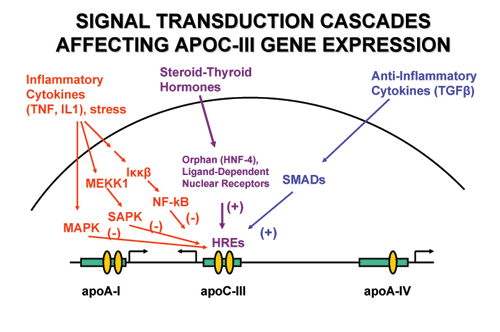
Signal transduction cascades triggered by pro- and anti- inflammatory
cytokines that affect positively or negatively the activity of hormone
nuclear receptors which bind to the HREs of the genes of the apoA-I/C-III/A-IV
gene cluster.
b) Transcriptional regulation of the human apoE gene in macrophages and
in the brain: Apolipoprotein E (apoE) is a key protein component of plasma
lipoproteins and promotes the clearance of excess cholesterol from the
plasma by binding to several members of the LDL receptor family. ApoE
deficiency in animal models and in human patients with type III hyperlipoproteinemia
is associated with premature atherosclerosis. ApoE has lately emerged
as an important molecule in several biological processes and diseases
such as cognitive function, immunoregulation, Alzheimer's disease (AD),
and even infectious diseases. ApoE expression is regulated by a complex
interaction of developmental, hormonal, dietary and pathological factors.
We are concentrating our efforts on the identification andcharacteriztion
of the regulatory elements responsible for the basal level of apoE expression
in the healthy state and for the modulation of its expression in pathological
states. The experiments are focused on two different cell types involved
in two distinct major diseases in which inflammation takes an important
counterpart: macrophages, one of the key cell types involved in atherogenesis
and astrocytes, one of the major cell types implicated in Alzheimers
Disease (AD).
The beneficial effect of apoE production by macrophages has been shown
in numerous animal studies: transgenic mice expressing apoE only in macrophages
are protected against atherosclerosis, even though the plasma levels of
apoE are exceedingly low and the animals are hypercholesterolemic; transgenic
mice with normal level of apoE expression except in macrophages are more
susceptible to atherosclerosis. On the other hand, it was demonstrated
that apoE plays a key role in neuronal repair after injury by redistributing
lipids to regenerating axons and to Schwann cells; also it has a protective
role for AD.
c) Transcriptional regulation of the lipid transporter ABCA1: Clinical
and epidemiological studies have shown an inverse correlation between
HDL cholesterol and the risk of coronary heart disease events. In fact,
low HDL cholesterol were found with high frequency in patients with premature
myocardial infarction. Formation of HDL requires functional interactions
of lipid-free apoA-I with the lipid transporter ABCA1. Specific mutations
in ABCA1 and/or deficiency in apoA-I prevent the formation of HDL. ABCA1
functions in the efflux of cellular cholesterol and phospholipids and
these functions are severly reduced in patients with Tangier disease that
have defective ABCA1 forms. In contrast, overexpression of ABCA1 is associated
with elevated plasma HDL levels, decreased plasma LDL levels and protection
from atherosclerosis. Selective increase in the expression of ABCA1 that
will ultimately result in increase in HDL requires a thorough understanding
of transcriptional regulatory mechanisms that control the expresison of
the ABCA1 gene.
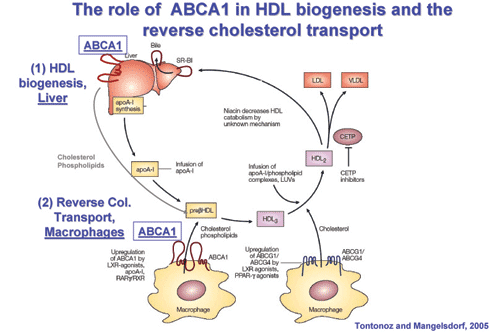
Schematic representation of the role of the ABCA1 lipid transporter in
HDL biogenesis and the reverse cholesterol transport.
Our aim is to understand the molecular mechanisms that control the hepatic
expression of the ABCA1 gene in vitro and in vivo. Specifically, we are
characterizing important regulatory regions present in the upstream ABCA1
promoter (class1/2 transcripts) as well as in the first intron, which
contains several transcription initiation sites for the class 3 transcripts
of this gene. We are also studying the role of Sp1 in the regulation of
ABCA1 gene expression by cholesterol via the Liver X Receptors (LXRs).
Finally, we are studying the mechanisms of differentiation-induced transacription
of the ABCA1 gene in macrophages.
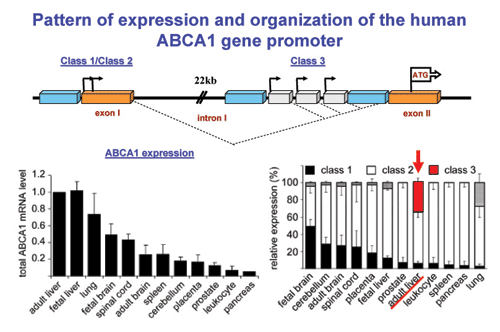
Patterns of expression and organization of the 5 region of the human
ABCA1 gene showing the sites of intitation of transcription of the Class1-3
transcripts.
|
